Measurement error in sibling models
Herasight's correction is correct
Emil Kirkegaard recently blogged on correcting for attenuation in sibling models. This was motivated by a recent Herasight intelligence polygenic score paper which apparently uses a correction found in Frisell et al 2012 (Sibling comparison designs: bias from non-shared confounders and measurement error). In his post, Kirkegaard says Frisell et al 2012’s correction is wrong. This inspired me to write my own version of it, along with a simulation, to see if I could improve the formula.
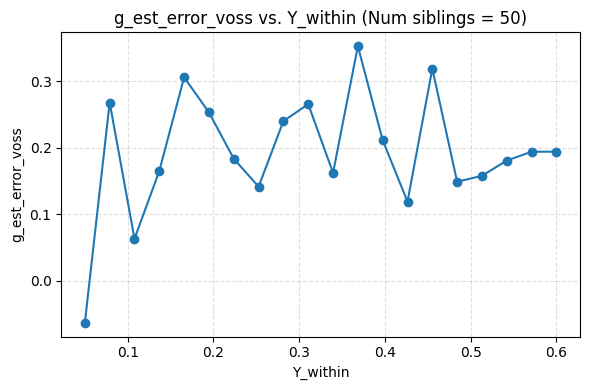
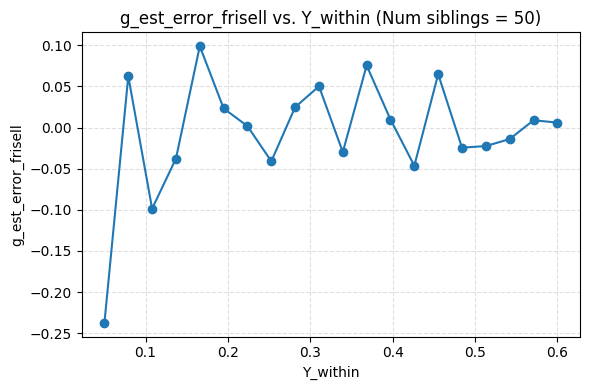
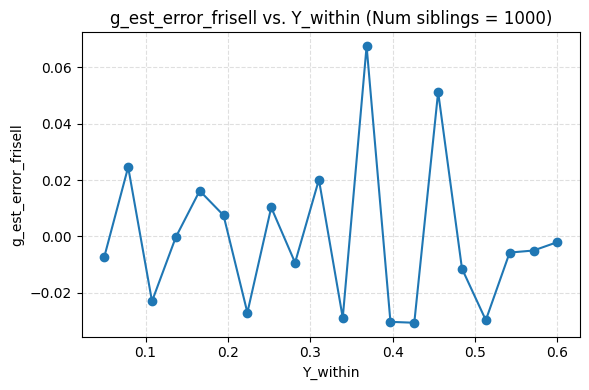
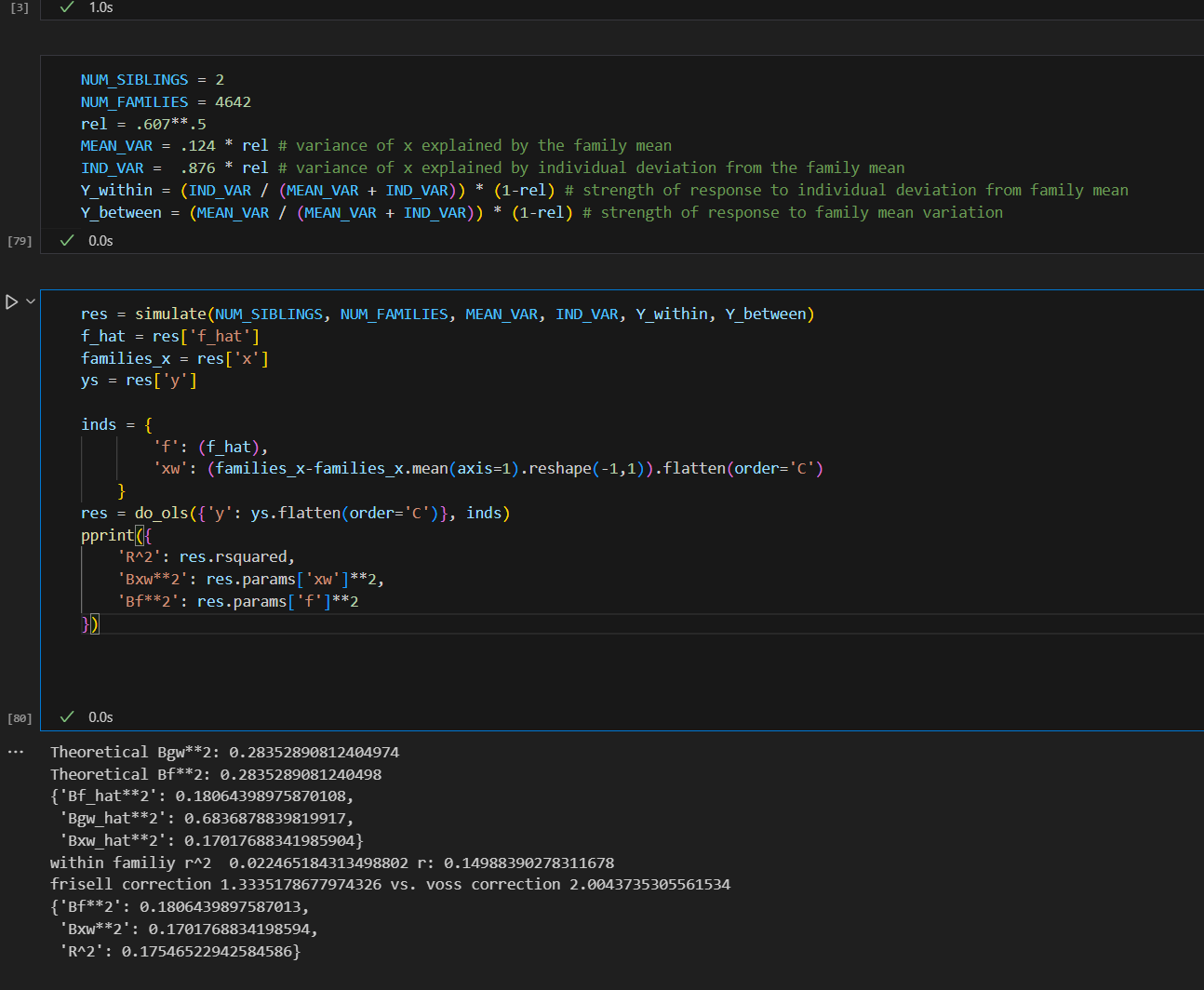
I ended up finding an alternative formula, but it yields the same results as Frisell 2012, which per my simulation actually produced a correct formula. Sadly, my formula is less efficient, i.e. it appears to produce noisier estimates. It is also only asymptotically unbiased in number of points per cluster, while theirs is unbiased. For now I’m just putting it up on github here.
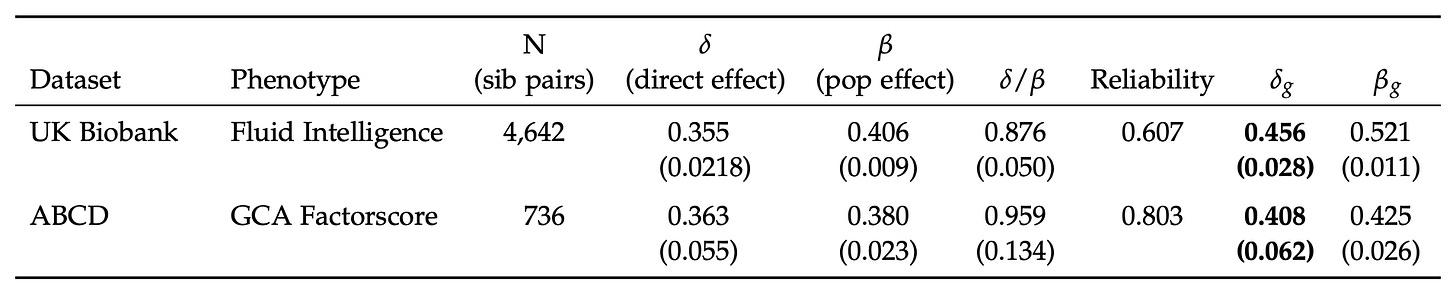
Plugging in the data from the Herasight paper for the UK Biobank, I found a Frisell corection of 1.33. It looks like they used about 1.28. Using 1.33 would give .472 instead of .456. The confidence intervals overlap enough that this doesn’t matter.
I’m quite confident their correction factor is not flawed in any practical way.


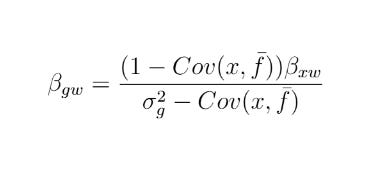
Mind making a response, he mentioned you
https://open.substack.com/pub/kossina/p/going-to-bat-for-the-age-of-consent?r=31ft4d&utm_campaign=post&utm_medium=web&showWelcomeOnShare=false
It depends on standardized vs not standardized variables.